PRIN2022 - Identification of genetic and epigenetic regulators supervising R-loops formation

Titolo progetto: PRIN2022 - Identification of genetic and epigenetic regulators supervising R-loops formation
Programma di finanziamento: PRIN 2022
Responsabile scientifico: dott. Eros Di Giorgio
Ruolo del DAME: partner
Descrizione generale:
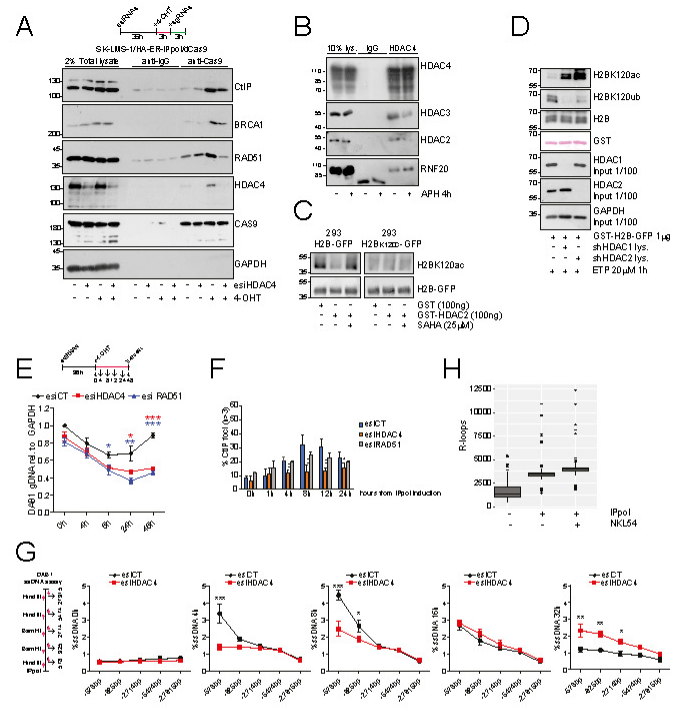
The study of the so called 4D genome has clarified that RNA molecules play a key role in organizing the DNA into High-Order Chromatin Structures (HOCS). In particular, transient and highly frequent DNA:RNA hybrids can stabilize somewhere in the genome by adopting a triple-stranded structures called R-loop. R-loops were identified as the result of abortive transcripts accumulation, but more recently it has been demonstrated that they actively regulate transcription by serving both as platforms for the recruitment of transcription factors and terminators. Moreover R-loops can also form in trans independently from the transcription process and affect genome stability and DNA damage response (DDR). In this proposal we aim to identify new cis and trans regulators of R-loops and evaluate the direct impact of Rloops accumulation on genome stability and DSB repair. In Task 1 we aim to describe a new epigenetic mechanism that controls R-loops metabolism. We have demonstrated that a newly identified HDAC complex, named HRDAC, supervises the acetylation of H2B at lysine 120 thus controlling R-loops formation, in particular in correspondence to genomic loci subjected to double-strand breaks (DSBs). Here by inhibiting or by knocking-down key HRDAC subunits we aim to correlate RNAP2 processivity to R-loops by means of DRIP-seq, DRIVE-seq and PRO-seq. Colorectal cancer (CRC) cell lines and Patients’ derived organoids (PDOs) will be engineered to express mega-endonucleases and DDR-reporters and quantify the impact of R-loops on the repair of well-defined DSBs. In Task 2 the topological stress and the replication/transcription (R/T) conflicts will be investigated as cis and trans sources of R-loops. Topoisomerase I inhibition (CPT) will be used as an exogenous inducer of topological and replication stress. DRIP-seq and TRIPn-Seq will be adopted to correlate R-loops formation to R/T conflicts. The genomic instability induced by R-loops accumulation will be quantified both at single cell level and by omics approaches. In Task 3, ATM/Tel1 signaling will be correlated to R-loops formation. Mutant yeast strains will be adopted to increase or decrease R-loops, while DRIP-seq and ChIP-seq will be used for the co-mapping of Tel1 and DNA:RNA hybrids. The levels of DNA damage after CPT treatment in cells lacking Tel1 and R-loop resolving factors will be evaluated and compared to wild-type strain. Finally, synthetic lethality will be exploited in Task 1 by combining FOLFOX therapy and epigenetic drugs on CRC PDOs. On the opposite, in Task 3 a yeast genetic screening will be adopted to identify compensatory pathways overcoming ATM/Tel1, Sen1 and RNAseH deficiency thus allowing survival in the presence of CPT and hydroxyurea. In conclusion, our project will identify new R-loops regulators; moreover, by quantifying the impact of R-loops on genome stability, we also aim to explore new approaches of personalized medicine for the treatment of genetic diseases and cancer.
Partner del progetto:
- Università di Bologna
- Università degli Studi di Udine
- Università degli Studi di Milano-Bicocca
Date inizio e fine progetto: 28.09.2023 – 27.09.2025
Budget totale del progetto: 76.230,00€
Sito web: https://prin.mur.gov.it/
Finanziato dall’Unione europea – Next Generation EU